An overview of neuroimaging file formats - from the user's perspective
At first, I found the plethora of file formats in neuroimaging quite overwhelming and with every processing software my confusion just seemed to increase exponentially. Despite the large number of resources online, I was missing a summary that describes the file formats in brevity but at the same time will answer the basic questions. I’m thinking of a typical user who doesn’t have a deep technical background in computer science, and who might already be stuck at the question: How can I actually open the file? (I’m exactly such a typical user..) So I started to collect notes during my PhD, which I compiled here as an overview ‘from the user’s perspective’. Of course, this list is not exhaustive - and it might even be outdated soon - but it covers the most commonly used filetypes for MRI-based neuroimaging.
My understanding of file formats and data storage in general expanded strongly when I had to manipulate files outside of the conventional processing software. That’s why I included some code snippets (code here) at the end that demonstrate of how files can be manually loaded and changed.
Volume-based data
Nifti
- What does it store?
- a rectangular matrix of image intensities from data that is organized in voxels
- typically 3D (3 spatial coordinates), but the 4th dimension can encode time in an fMRI scan
- examples: a brain scan, manually drawn ROIs, diffusion tensors, a non-linear registration field
- Acronym:
- Neuroimaging Informations Technology Initiative
- filename extension:
- nii
- nii.gz, if compressed
- General information:
- Nifti is by far the most commonly used file format to store volumetric brain imaging data and most processing softwares will support it. It originated from the ANALYZE 7.5 format and a Nifti file stores both the data matrix and a header that contains meta data. Due to the large file size, Nifti files are often stored compressed using gzip (nii.gz), which performs lossless compression based on the DEFLATE algorithm. There is a Nifti1 and a Nifti2 file format, but usually we don’t have to worry about that.
- How can I display it?
- There are various viewers available, just to name a few: FSLeyes, freeview, mango, wb_view, MRIcro, Brainstorm
- How can I manipulate it?
- In general, brainimaging softwares come with their own functions to perform analysis on Nifti files or perform basic manipulations
- Processing softwares are, for example: FSL, SPM, AFNI, Freesurfer, DIPY
- Read raw file for manual manipulation:
- matlab: Niftiread
- Python: nibabel Nifti module
Dicom
- What does it store?
- Raw brain scan data that comes directly from the MRI scanner
- (raw means, here the data has been transformed from k-space to voxel space)
- each 2D slice of the brain is stored in a separate file
- Acronym:
- Digital Imaging and Communications in Medicine
- filename extension:
- usually: none (the file type is inferred by a characters in a specific location of the header)
- sometimes: .dcm
- General Information
- The DICOM format is widely used in medial imaging, not only for brain scans and has to serve many purposes
- The file header can contain complex information that might not be needed for neuroimaging analysis
- How can I display it?
- Many viewers from the medical context, but also FSLeyes, Mango, Brainstorm
- How can I manipulate it?
- Typically, you don’t want to manipulate a DICOM file directly
- Conversion to Nifti for example with dcm2nii from MRIcro tool
- Read raw file for manual manipulation:
- MATLAB: dicomread
- Python (limited support): pydicom, which is used as back-end for reading DICOMs in nibabel
mgh
- What does it store?
- see above for Nifti
- Acronym:
- Massachusetts General Hospital
- filename extension:
- mgh
- mgz (if compressed with ZLib)
- General information:
- The format is specifically used within the Freesurfer framework and has similar functionality and properties as the Nifti format
- How can I display it?
- The native fiewer is freeview, but also FSLeyes
- How can I manipulate it?
- Native processing tools are Freesurfer commands
- convert to Nifti using the Freesurfer command mri_convert
- Read raw file for manual manipulation:
- MATLAB: MRIread
- Python: nibabel, freesurfer.io
minc
- What does it store?
- see above for Nifti
- Acronym:
- Medical Image NetCDF
- filename extension:
- .mnc
- General Information:
- The file format was developed at the MNI and it is designed to work with the the MINC processing toolbox and viewer
- There is a MINC1 and a MINC2 file format
- How can I display it?
- The native viewer is Display, but also: FSLeyes, freeview, Mango, Brainstorm, Register
- How can I manipulate it?
- Native processing tools is the Minc toolbox, but also Freesufer, AFNI
- Read raw file for manual manipulation:
- Matlab: loadminc
- Python: nibabel, preliminary MINC2 support
Suface-based data
Gifti
- What does it store?
- surface-based data that is organized by vertices
- can store Surface geometry and vertex-wise data
- Nifti for surfaces
- examples: a pial surface mesh, a surface map of cortical myelin, a manually drawn surface ROI
- Acronym:
- Geometry Informatics Technology Initiative
- filename extension:
- .gii
- two-part file extension depending on datatype
- Surface geometry files (.surf.gii)
- metric files (.func.gii, .shape.gii)
- label files (.label.gii)
- General information:
- geometry data (surf.gii) represents a surface mesh. The files store the 3D coordinates and triangle arrays of a surface
- Metric files (func.gii and shape.gii, there is no difference) store continuous values that are associated with the vertices. The data is a one-dimensional vector of intensities.
- Label data (label.gii) stores integer values for each vertex together with a name and a colour
- The file organization is based on the XML format
- The format is widely used within the Human Connectome Project
- How can I display it?
- wb_view, freeview, FSLeyes, mango, Brainstorm
- metric and label files need to be loaded together with a surf.gii file that defines the spatial configuration
- How can I manipulate it?
- wb command line tools, AFNI tools, FSL, Freesurfer
- convert between file formats using mris_convert
- Read raw file for manual manipulation:
- Matlab: Gifti library
- Python: nibabel Gifti module
Cifti
- What does it store?
- data organized in ‘brainordinates’
- elements are cortical surface vertices and subcortical voxels
- example: dense connectome of cortical surface and subcortical structures
- Acronym:
- Connectivity format of the Geometry Informatics Technology Initiative
- filename extension:
- .nii (typically not compressed)
- two-part file ending for different datatypes:
- timeseries (dtseries.nii)
- parcellation (dlabel.nii)
- scalars (dscalar.nii)
- connectivity (dconn.nii)
- General information:
- The Cifti format was designed to handle data from disjoint structures, such as cortex and subcortical structures, and large connectivity matrices
- all relevant elements, i.e. all brainordinates could not be represented in a Nifti file alone
- Cifti is based on the Nifti, but it can include both volumetric and surface data
- The file organization is based on the XML format, which stores data arrays
- in addition to the data array, we store the ‘mapping’, which interprets the element’s index, i.e. it assigns the ‘position’ of each element
- mapping types can assign for examples parcels, or labels
- How can I display it?
- wb_view, freeview
- Cifti files need to be loaded together with a surf.gii file that defines the spatial configuration
- How can I manipulate it?
- workbench command line tools, FSL
- Read raw file for manual manipulation:
- MATLB: Cifti-matlab toolbox
- Python: nibabel: Cifti2 module
Miscellaneous data
wb_view-specific:
- When working with files related to HCP data, you will encounter additional file types that have been customized for the wb_view software:
- .scene and .spec files: organize which files are loaded an how they are displayed
- .border, .foci, .annot: additional features on the brain surface
Freesurfer-specific:
- some Freesurfer binary files have no filename extension and are only recognized by native commands
- gca, bshort, bfloat, COR, surface, curv, w, annot, patch, gcs, dat, xfm, m3d and lta
json
- json files store attribute/value pairs, which can hold additional non-primary data such as parameters and timing information. Json files are recommended in the BIDS format to store meta information related to primary imaging and behavioural data files.
Code snippets
Manipulate a Nifti file
This is a basic example of how, to load, modify and save a Nifti-file using Python’s nibabel library. The file should like like this before and after the manipulation:
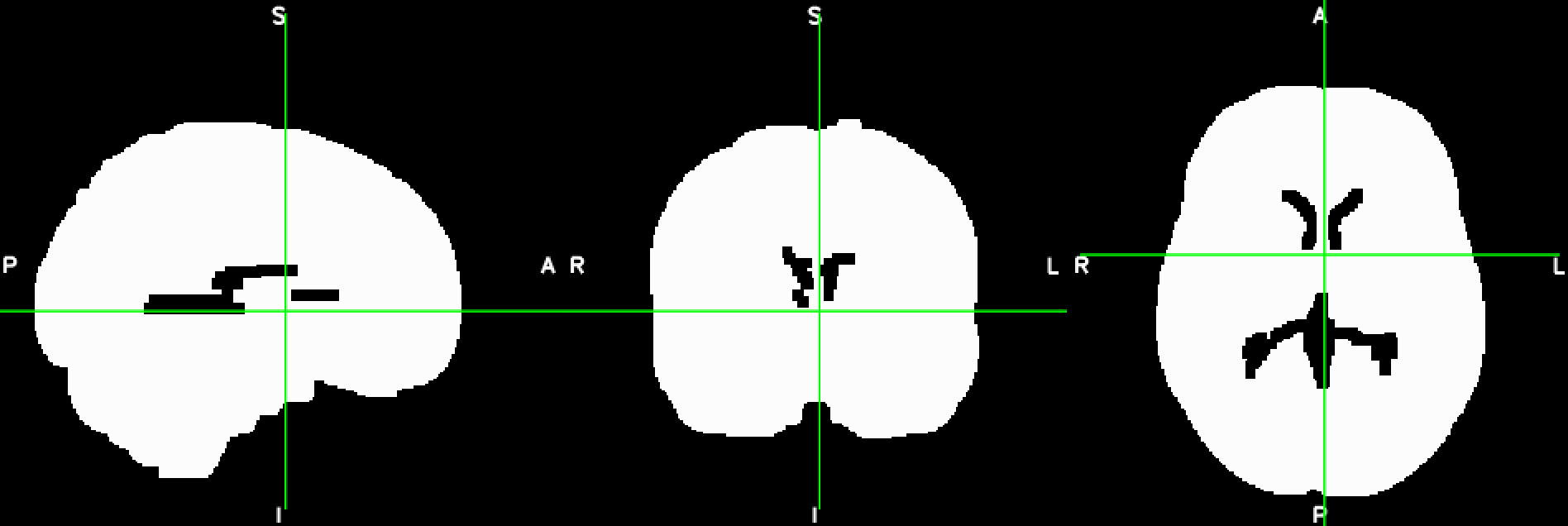
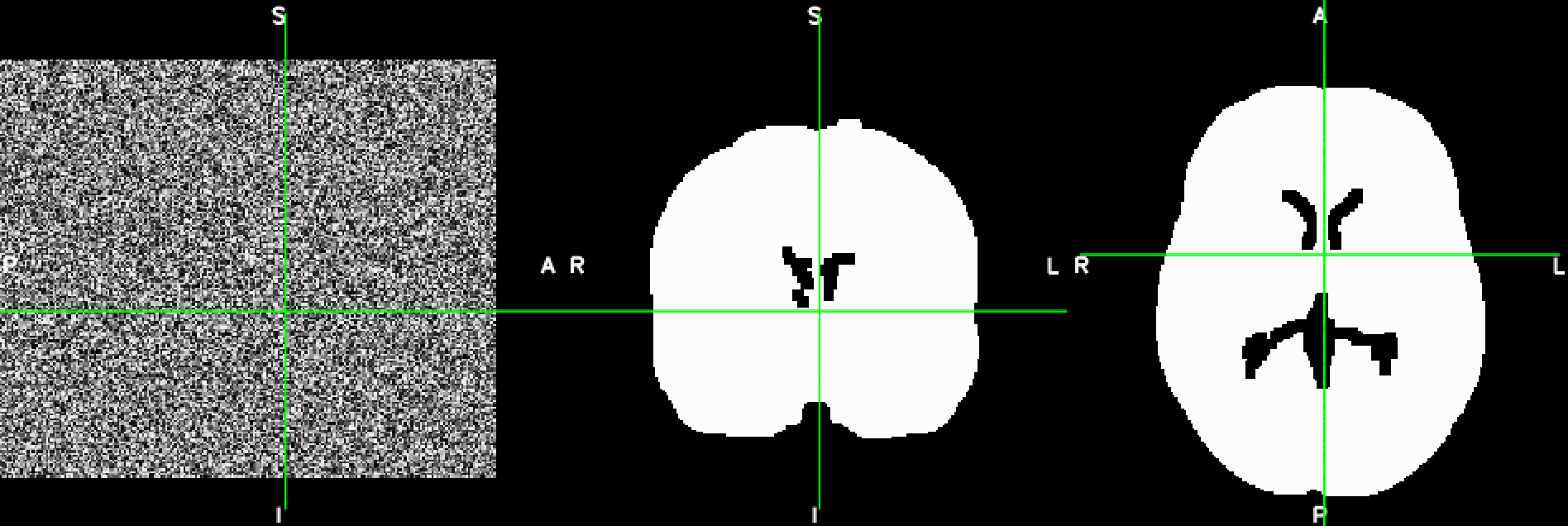
import numpy as np
import nibabel as nib
# load Nifti image
img = nib.load('example_brain.nii.gz')
# get data matrix
img_data = img.get_data()
# create copy of data matrix which you can modify
new_data = img_data.copy()
# fill one slice in the middle of the first axis (i.e. the mid-saggital slice) with random values
mid_index = np.int(np.rint(new_data.shape[0]/2))
new_data[mid_index, :, :] = np.random.rand(new_data.shape[1], new_data.shape[2])
# create new image object with header information of original image
new_img = nib.Nifti1Image(new_data, img.affine, img.header)
# save file
nib.save(new_img,'example_brain_modified.nii.gz')
Manipulate a border file
I didn’t include an example of how to manipulate a Gifti-file, because it works very similar as for the Nifti file. Modifying a wb_view border file, is more tricky, because it’s not supported by the nibabel library. In the example below I’m using the build-in python XML parser to load a border file to edit the colour associated with both the border class and the border name. Note that you can only display the border file together with geometric gifti file, in this case a sphere.
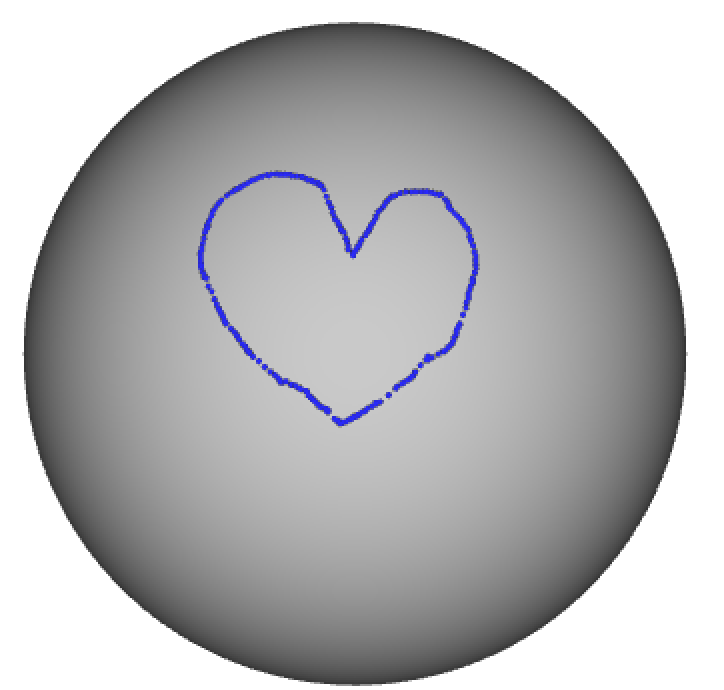
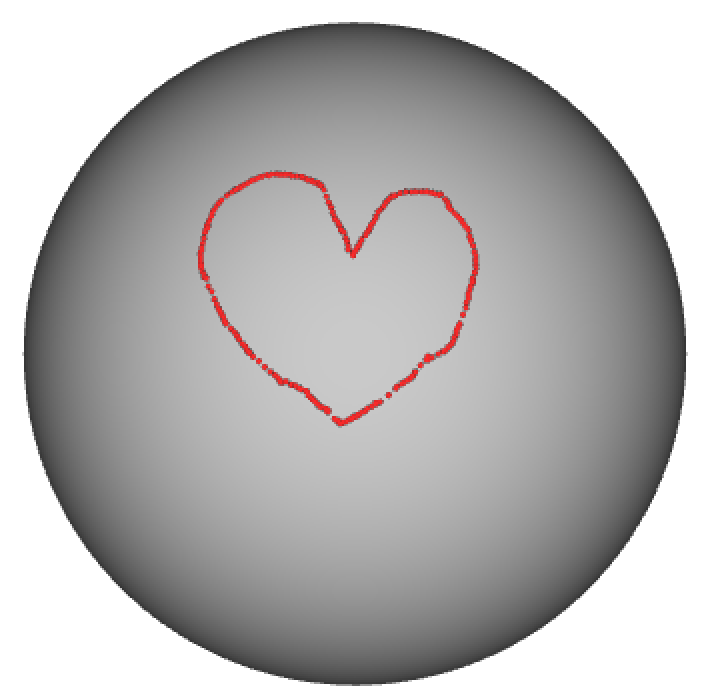
import xml.etree.ElementTree as ET
import numpy as np
# read in border file using XML parser
tree = ET.parse('example_border.border')
root = tree.getroot()
# access the top-level border_class and update colour
border_class = root.getchildren()[0]
border_class.set('Red','1')
border_class.set('Green','0')
border_class.set('Blue','0')
# access the low-level border_name and update colour
border_name = border_class.getchildren()[0]
border_name.set('Red','1')
border_name.set('Green','0')
border_name.set('Blue','0')
# save changes to new file
tree.write('example_border_red.border')
That’s it!
Thanks for reading this post :-)
Nicole